Deciphering splicing regulation
Work from our lab and others has shown causal roles for splicing factor dysregulation in tumors, yet what triggers changes in splicing factors in tumors remains incompletely understood. In solid tumors, mutations in splicing factor genes are rare (1-3% of patients), and therefore defining which molecular mechanisms lead to splicing factor dysregulation is greatly needed. These fundamental discoveries provide novel mechanisms that can be leveraged in the future to target the splicing machinery in disease.
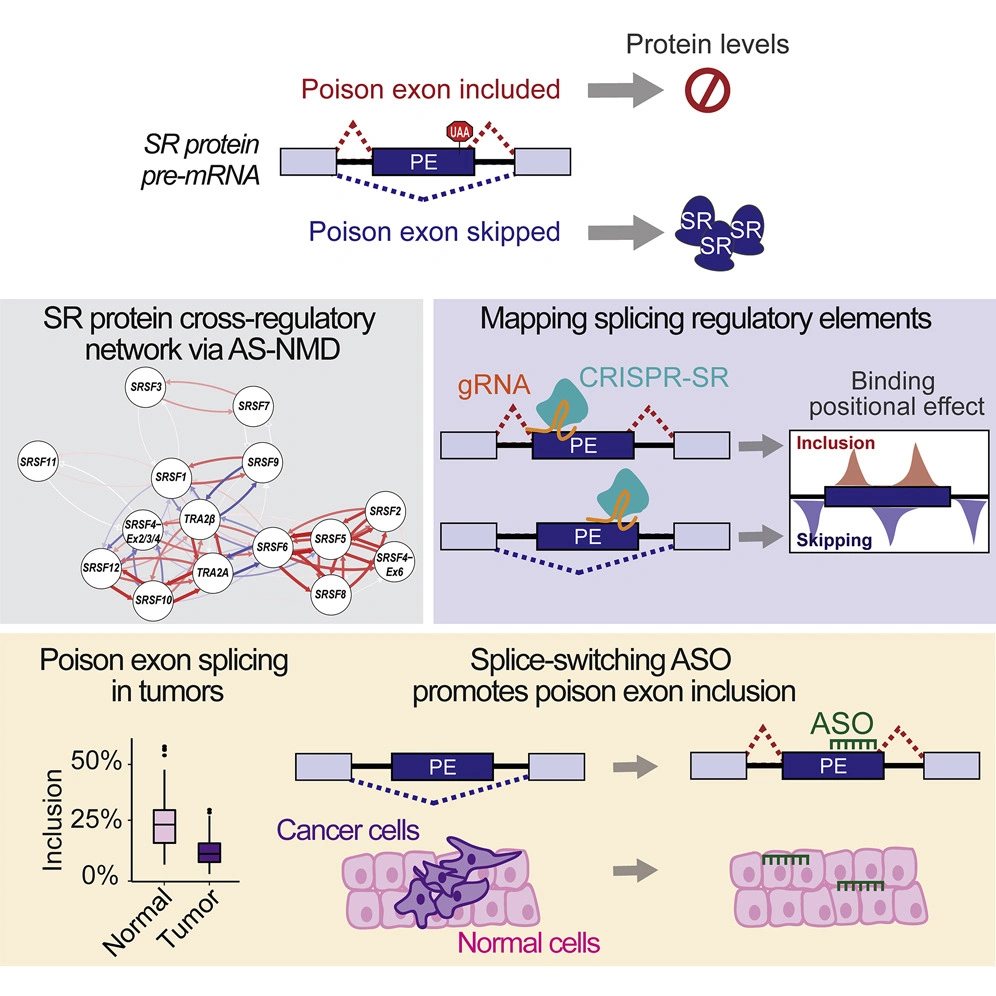
Our lab uncovered that splicing factor alterations often arise due to changes in transcriptional and post-transcriptional regulatory pathways in tumors. Specifically, we uncovered a MYC-regulated splicing network conserved across multiple tumor types, and a pan-cancer splicing signature of MYC activity associated with patient’s survival. In parallel, we revealed a novel mechanism by which splicing factors levels are coordinated via splicing of ultra conserved non-coding poison exons in normal cells. Our work reveals that changes in poison exon inclusion contributes to splicing factor dysregulation in disease.
Given the fact that aging is the greatest risk factor for most cancers, our lab has been systematically characterizing the aging-related splicing landscape, how it is regulated in cancer-initiating cells and their environment, and identified aged-driven pro-tumorigenic isoforms. Our work revealed how aging rewires the cellular composition of mammary tissues and impacts the transcriptomic and epigenomic programs of mammary epithelial, fibroblast, and immune cells, identifying shared signatures of aging and cancer. We have also uncovered age-related changes in the splicing repertoire and splicing factor levels in both human and mouse mammary tissues, some of which are also found in human breast tumors. Finally, we uncovered age-related chromatin accessibility changes that could affect RNA splicing through both cis and trans mechanisms.
We are now defining how age-associated epigenetic and alternative splicing alterations contribute to breast cancer initiation through an integrated multi-omics mouse-human approach. These mechanistic discoveries into the biology of cancer initiation are expected to lead to new approaches for early detection, intervention, and prevention for the most common form of cancer in women.
Relevant publications:
- Angarola BL*, Sharma S*, Katiyar N, Kang HG, Nehar-Belaid D, Park S, Gott R, Eryilmaz GN, LaBarge M, Palucka K, Chuang JH, Korstanje R, Ucar D#, Anczuków O# (2024). Comprehensive single cell aging atlas of mammary tissues reveals shared epigenomic and transcriptomic signatures of aging and cancer. Nature Aging, in press. *equal contribution. #Corresponding author.
- Urbanski L, Brugiolo M, Park S, Angarola BL, Leclair NK, Palmer P, Keshari Sahu S, Anczuków O# (2022). MYC regulates a pan-cancer network of co-expressed oncogenic splicing factors. Cell Reports 41(8), 11704. PMC9731204. #Corresponding author.
- Leclair NK, Brugiolo M, Urbanski L, Lawson S, Thakar K, Yurieva M, George J, Hinson JT, Cheng A, Graveley BR, Anczuków O# (2020). Poison exon splicing regulates a coordinated network of SR protein expression during differentiation and tumorigenesis. Molecular Cell, 80(4):648-665.e9. PMC7680420. #Corresponding author.
- Park S*, Brugiolo M*, Akerman M*, Das S*, Urbanski L, Geier A, Kesarwani AK, Fan M, Leclair NK, Lin KT, Hua L, Hu I, George J, Muthuswamy SK, Krainer AR#, Anczukow O# (2019). Differential functions of splicing factors in mammary transformation and breast cancer metastasis. Cell Reports, 29(9), 2672-2688.e7. PMC6936330. *equal contribution. #Corresponding author.
Targeting RNA splicing in disease
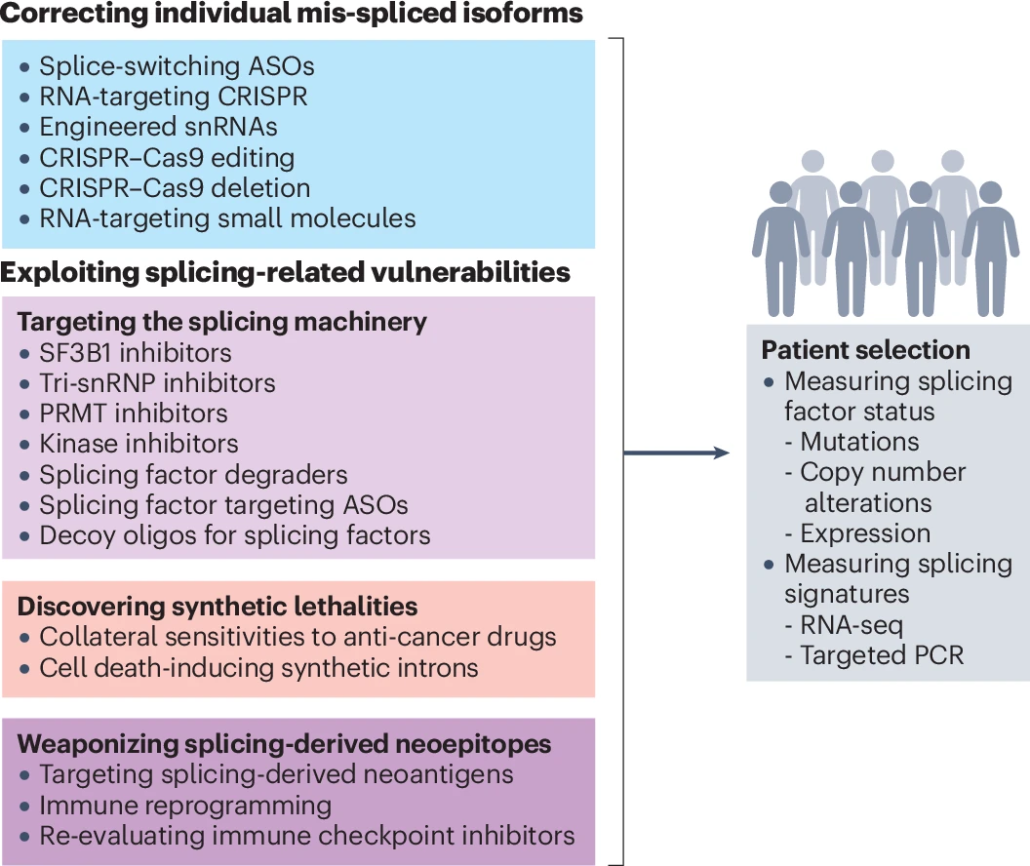
Most splicing factors are difficult to target using small molecules due to the lack of a catalytic site and their essential roles in normal cells. Our work has produced two novel RNA-based approaches to target splicing factors in cancer models.
First, we have adapted an RNA-targeting CRISPR system to guide an artificial splicing factor to a given intronic or exonic location and promote exon inclusion or skipping in a position-dependent manner. This flexible and scalable system can be used to interrogate the functions of specific splicing factors and define the rules for their activity, as well as to correct aberrant splicing of selected pathogenic isoforms.
In parallel, leveraging our discovery of the regulatory role of poison exons, we developed approaches to increase poison exon-inclusion and therefore decrease splicing factor protein levels in preclinical models. This strategy is widely applicable across tumor types to hundreds of other splicing factors that are regulated by similar mechanisms for which no specific inhibitors exist to date. This means that technology platforms broadly applicable across fields now exist, which, in combination with our mechanistic discoveries, can be applied to target cancer-causing splicing alterations.
We are now developing splice-switching approaches that simultaneously probe the biology of a specific isoform and define its regulators and regulatory elements. These tools will enable testing the pathogenicity of any given spliced isoform using in vitro or in vivo models of human diseases with splicing alterations. We are also leveraging our mechanistic insights into the regulation of splicing factors in aging and cancer to develop novel RNA-based approaches for modulating the levels and activity of specific splicing factors.
Relevant publications:
- Leclair NK, Chen WC, Zakimi Naomi, Choudhury A, Magill S, Shen E, Bulsara KR, Raleigh D#, Anczuków O# (2024). RNA splicing as a biomarker and phenotypic driver of meningioma DNA methylation groups. Neuro-oncology, Aug 2:noae150. #Corresponding author.
- Leclair NK, Brugiolo M, Urbanski L, Lawson S, Thakar K, Yurieva M, George J, Hinson JT, Cheng A, Graveley BR, Anczuków O# (2020). Poison exon splicing regulates a coordinated network of SR protein expression during differentiation and tumorigenesis. Molecular Cell, 80(4):648-665.e9. PMC7680420. #Corresponding author.
- Modified Antisense Oligonucleotides Targeting Splicing Factors. Anczukow O, Leclair N. US 63/110,225 (11/05/2020).
- Compositions And Methods For Treating Meningioma. Anczukow O, Leclair N. US 63/685,365 (08/21/2024)
Our collaborators
We believe collaboration and team science are essential drivers of innovation and discovery in today’s complex research landscape. By bringing together diverse perspectives, skills, and expertise, our collaborative programs enable us to tackle multifaceted problems that cannot be addressed by any one lab alone. We currently have ongoing collaborative research projects with: Duygu Ucar (Jax), Karolina Palucka (Jax), Ron Korsantje (Jax), Jeffrey Chuang (Jax), Christine Beck (Jax), Charles Lee (Jax), Mark Labarge (COH), David Raleigh (UCSF), Harish Vadudevan (UCSF), and others.